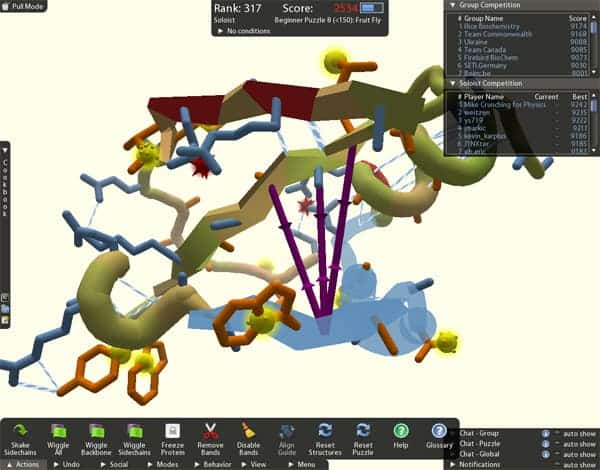
Scientists from the University of Washington have been struggling for the past decade to decipher the complex structure of an enzyme that exhibits behavior similar to that of an enzyme key in the development of AIDS from an HIV infection, and which might hold a critical role in building a cure for the disease. Gamers playing spatial game Foldit have managed to collectively determine the enzyme’s structure in ten days.
Puzzled by the intricate structure of the M-PMV retroviral protein, an enzyme that plays a key role in the development of a virus similar to HIV, scientists have striven to find its chemical key for ten years now. Each enzyme has millions of possible combination in which it can fold its atom bonds, and determining its precise structure is a very laborious enterprise even for high-end computers with large processing power.
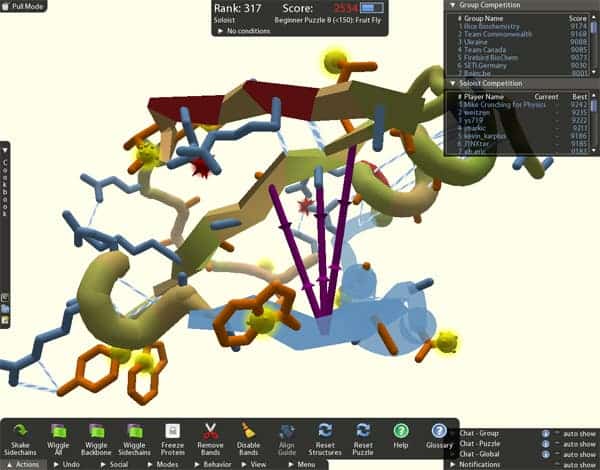
As a long-shot University of Washington biologists sent the virtual 3D model of the M-PMV to the online game Foldit, where gamers folded and turned it into a myriad of combinations. Eventually, and remarkably enough, the gamers obtained the optimum one – the state that needed the lowest energy to maintain. Now unlocked, scientists have a concrete means of understanding how the enzyme works, and consequently how to attack it.
“This was really kind of a last-ditch effort. Can the Foldit players really solve it?” Firas Khatib, a biochemist at the University of Washington and the lead author on the recently published research paper told MSNBC. “They actually did it in less than 10 days.”
Foldit is a very simple game, which tackles biology’s biggest issue – folding proteins. To play the game you don’t need any biology background, just your native spatial reasoning skills. Motivation comes in the form of competition, and from this stand point, the game has been more than suitably designed. Basically you get scored for three factors: how well you pack the protein, how efficiently you hide the hydrophobics and how you clear the clashes. Trust me, it’s a lot simpler than it sounds.
“Foldit attempts to predict the structure of a protein by taking advantage of humans’ puzzle-solving intuitions and having people play competitively to fold the best proteins,” states the game’s website.
“Since proteins are part of so many diseases, they can also be part of the cure.
Protein folding had proved to be one of the more popular uses for distributed computing
“Players can design brand new proteins that could help prevent or treat important diseases.”
The game allows players to chat with each other and collaborate, thus various gamers built up each others work and collectively managed to crack the code for the most energy efficient enzyme structure – the most important structure to study.
The reason why computers haven’t been able to do this, despite their evidently superior processing capabilities, is that they’re still far from being capable of having human-like spatial reasoning. Interestingly enough, Foldit records the players’ actions and processes them in an algorithm which will eventually help the AI behind the game to someday be able to compile successful structures on its own.
Seth Cooper, a University of Washington computer scientist and lead designer and developer of Foldit, is hoping that more scientists send them problems that fit within the Foldit format.
“The critical role of Foldit players in the solution of the M-PMV [retroviral protease] structure shows the power of online games to channel human intuition and three-dimensional pattern-matching skills to solve challenging scientific problems,” said the study, which was published by Nature Structural & Molecular Biology. “Although much attention has recently been given to the potential of crowdsourcing and game playing, this is the first instance that we are aware of in which online gamers solved a longstanding scientific problem.”
Still, the breakthrough is amazing by all means. Next time somebody tells you you’re wasting time playing a video game, you can always show them this article and tell them you’re helping save the world.
Article revised 10/16/2013: 1. In the original draft, “Washington University” was mistakenly written instead of “University of Washington”
2. Grammar fix
3. Initial paragraph was modified, better reflecting reality.